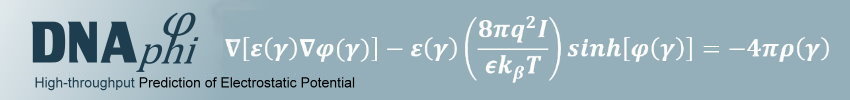About DNAphi
 Mechanisms of protein-DNA recognition are at the core of transcriptional regulation,
and it has become clear that the readout of DNA shape is an important component of the DNA binding specificity of transcription factors.
While methods for the high-throughput prediction of DNA shape features have been developed
(DNAshape),
minor groove width has often just been a "proxy" for electrostatic potential,
in part because it was not possible to calculate electrostatic potential for thousands of sequences or entire genomes.
A/T and G/C base pairs carry different functional groups in the minor groove that affect electrostatic potential through their steric impact on groove geometry in addition to their different charges.
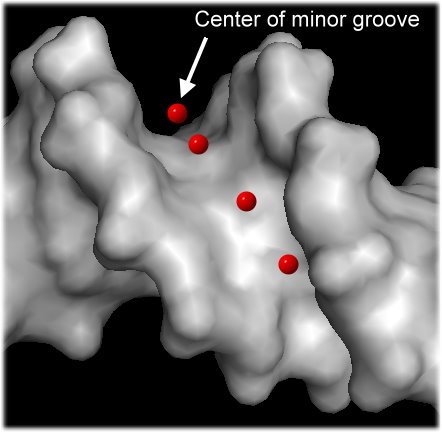
To capture both effects and thus physical interactions, we developed a new method to calculate the electrostatic potential "phi" in the minor groove (see figure) in a high-throughput manner for any length or number of sequences based on the data mining of results from solving the non-linear Poisson-Boltzmann equation for many DNA fragments with diverse sequences
(Go to φ prediction!).
To model DNA binding specificities of transcription factor using electrostatic potential, we included a statistical machine learning approach (MLR) for quantitative prediction of DNA binding specificities of transcription factors
(Go to φ learning!).
Mechanisms of protein-DNA recognition are at the core of transcriptional regulation,
and it has become clear that the readout of DNA shape is an important component of the DNA binding specificity of transcription factors.
While methods for the high-throughput prediction of DNA shape features have been developed
(DNAshape),
minor groove width has often just been a "proxy" for electrostatic potential,
in part because it was not possible to calculate electrostatic potential for thousands of sequences or entire genomes.
A/T and G/C base pairs carry different functional groups in the minor groove that affect electrostatic potential through their steric impact on groove geometry in addition to their different charges.
To capture both effects and thus physical interactions, we developed a new method to calculate the electrostatic potential "phi" in the minor groove (see figure) in a high-throughput manner for any length or number of sequences based on the data mining of results from solving the non-linear Poisson-Boltzmann equation for many DNA fragments with diverse sequences
(Go to φ prediction!).
To model DNA binding specificities of transcription factor using electrostatic potential, we included a statistical machine learning approach (MLR) for quantitative prediction of DNA binding specificities of transcription factors
(Go to φ learning!).
To cite DNAphi:
T.P. Chiu, S. Rao, R.S. Mann, B. Honig, R. Rohs: Genome-wide prediction of minor-groove electrostatic potential enables biophysical modeling of protein-DNA binding, Nucleic Acids Res. gkx915, https://doi.org/10.1093/nar/gkx915.